GSEA 介绍
GSEA全名Gene Set Enrichment Analysis,翻译过来就是基因集富集分析。基本思想是使用预定义的基因集,将基因按照在两类样本中的差异表达程度排序,然后检验预先设定的基因集是否在这个排序表的顶端或者末端富集。GSEA分析检测基因集合而不是单个基因的表达变化,因此可以包含更多细微的表达变化,从另一个角度来解读生物学信息,以阐述其中的生物学意义。
GSEA与常规富集分析
常规的富集分析(GO和KEGG)侧重于比较两组间差异基因,而容易遗漏掉部分差异不显著但是有生物学意义的基因,容易忽略一些基因的功能,生物特性,调控网络等有价值的信息。GSEA不需要特意选择差异基因,算法是根据其整体的表达趋势,找出基因中差异不明显但是整体趋势一致的基因集。
GSEA的原理
GSEA的假设是设定好的功能基因集里面的成员在目标基因集中是随机分布的。如果功能基因集是处于目标基因集的顶部或底部,且置换检验后P值显著,那么说明此功能基因集所表现的功能的实验或其他处理下发生了作用,也是需要关注的基因集。
GSEA中有个重要的概念Enrichment Score(ES,富集得分),它表示基因集成员在目标基因集中的富集程度。ES的具体算法如下图所示!
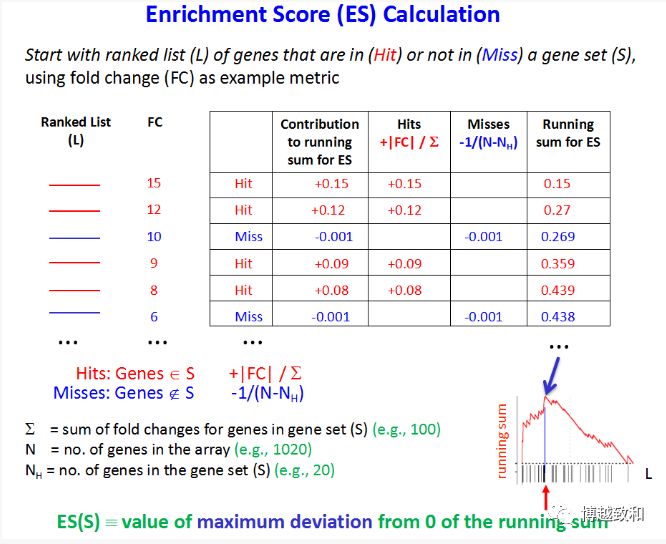
是指功能基因集
L指基因按Signal2Noise或fold change从大到小排序后的目标基因集
Ranked List红色代表基因位于功能基因集中,蓝色代表不在功能基因集中,如果在加分,不在减分
评估ES的显著性,通过permutation test来检验,转化不同分数下的数据或者做基因集的转化来重新计算ES,或ES分布有显著变化,那么说明此基因集是有生物学意义的
GSEA分析实战
方法一
使用Y叔所写的神包clusterProfiler,好像大家都已经很熟悉了这个R包,不仅能做Go和kegg的富集,还能做GSEA,包括GSEA Go,和GSEA Kegg,而且最重要的是使用非常简单,就一个函数就能做完分析。
在开始之前先导入需要用到的R包
library(data.table)
library(tidyverse)
library(magrittr)
library(knitr)
library(clusterProfiler)
library(enrichplot)
废话不多说,我们来准备GSEA的输入文件!
一.geneList
geneList 有三个特性
数值向量,可以是fold change或其他的数值组成
命名,数值向量的每个数值都有一个对应的基因名
排序,一般是数值从高到低排序的 比如使用DESeq2做完差异分析后,会得到如下表格。
## id baseMean log2FoldChange lfcSE stat pvalue padj
## 1 IQCG 493.95026 -3.164708 0.4352913 -7.270323 3.586299e-13 1.873303e-08
## 2 PABPC1L2B-AS1 20.35630 -4.337893 0.9577222 -4.529385 5.915550e-06 1.544994e-01
## 3 PTPRK 793.30041 -1.690615 0.3889629 -4.346469 1.383468e-05 2.408848e-01
## 4 MAGED4 25.76054 4.682533 1.1326350 4.134195 3.562017e-05 4.651549e-01
## 5 BX890604.2 165.29529 1.808737 0.4475756 4.041187 5.318128e-05 5.555848e-01
## 6 FP700111.2 65.29497 -2.190573 0.5589554 -3.919047 8.889973e-05 7.739462e-01
## 7 DMRTA2 264.05147 1.258098 0.3342768 3.763641 1.674576e-04 9.486864e-01
## 8 CXorf49 21.76600 5.682893 1.5173600 3.745250 1.802142e-04 9.486864e-01
## 9 VAC14-AS1 36.58607 -2.631516 0.7044571 -3.735523 1.873252e-04 9.486864e-01
## 10 ADGRA1-AS1 183.03451 1.608487 0.4315921 3.726870 1.938726e-04 9.486864e-01
## 11 SLX1B 13.18897 -6.497998 1.7561470 -3.700145 2.154767e-04 9.486864e-01
## 12 SLC20A1P2 12.76897 -4.918401 1.3480245 -3.648599 2.636740e-04 9.486864e-01
按fc排序得到gene_List
data$FC2
gene_List
names(gene_List)
gene_List
print(head(gene_List))
## FAM243B SLX1B OSTCP2 AC023866.2 HIST1H4L FABP5P11
## 42.95388 42.22397 40.96669 40.51999 39.24414 38.37089
二.功能基因集合
这里使用clusterProfiler自带的基因集文件作为测试基因集
gmtfile "extdata", "c5.cc.v5.0.entrez.gmt", package="clusterProfiler")
gene_set
print(head(gene_set))
## ont gene
## 1 NUCLEOPLASM 3190
## 2 NUCLEOPLASM 2547
## 3 NUCLEOPLASM 26173
## 4 NUCLEOPLASM 9439
## 5 NUCLEOPLASM 57508
## 6 NUCLEOPLASM 6837
可以看到基因ID使用的是EntrezID,为了与目标基因集保持一致,我们将目标基因集IDSYMBOL转化为EntrezID
library(org.Hs.eg.db)
gene_map"SYMBOL",toType = "ENTREZID",org.Hs.eg.db)
gene_map
print(head(gene_map))
## SYMBOL ENTREZID
## 1 FAM243B 102723451
## 2 SLX1B 79008
## 3 OSTCP2 646567
## 4 HIST1H4L 8368
## 5 FABP5P11 266699
## 6 RNY1P2 100862666
gene_List.entrez
names(gene_List.entrez)2]
gene_List.entrezis.na(names(gene_List.entrez))]
print(head(gene_List.entrez))
## 102723451 79008 646567 8368 266699 100862666
## 42.95388 42.22397 40.96669 40.51999 39.24414 38.37089
似乎一切都准备就绪,下面就开始进行GSEA分析,真就只需要一条命令就够了
gsea.raw1000,pvalueCutoff = 1,TERM2GENE=gene_set)
print(head(gsea.raw@result))
## ID Description setSize enrichmentScore NES pvalue p.adjust qvalues rank leading_edge core_enrichment
## FOCAL_ADHESION FOCAL_ADHESION FOCAL_ADHESION 13 -0.4466639 -1.401630 0.04545455 1 1 18408 tags=92%, list=55%, signal=41% 51466/51474/9564/7414/5788/87/143903/88/83543/7205/89
## TRANS_GOLGI_NETWORK_TRANSPORT_VESICLE TRANS_GOLGI_NETWORK_TRANSPORT_VESICLE TRANS_GOLGI_NETWORK_TRANSPORT_VESICLE 13 -0.6847663 -2.148795 0.04545455 1 1 10493 tags=100%, list=32%, signal=68% 9182/8906/26119/54812/261729/538/5045/51324/57120/11276/3482/1174
## SYNAPTIC_VESICLE SYNAPTIC_VESICLE SYNAPTIC_VESICLE 14 -0.4498060 -1.475309 0.05555556 1 1 18304 tags=100%, list=55%, signal=45% 55530/3382/3060/4504/273/23312/5538/23557/1201/51399/320/5864/8224
## CELL_SUBSTRATE_ADHERENS_JUNCTION CELL_SUBSTRATE_ADHERENS_JUNCTION CELL_SUBSTRATE_ADHERENS_JUNCTION 16 -0.4467042 -1.569570 0.07692308 1 1 18408 tags=94%, list=55%, signal=42% 51466/51474/9564/7414/4892/5788/87/10580/143903/88/83543/91624/7205/89
## LATE_ENDOSOME LATE_ENDOSOME LATE_ENDOSOME 12 -0.4373252 -1.333150 0.10714286 1 1 18718 tags=100%, list=56%, signal=44% 7879/23392/7251/51552/973/540/538/1497/7178/9367/3916
## CELL_MATRIX_JUNCTION CELL_MATRIX_JUNCTION CELL_MATRIX_JUNCTION 18 -0.4473638 -1.448694 0.11111111 1 1 13849 tags=94%, list=42%, signal=55% 51466/51474/667/9564/7414/4892/5788/87/10580/55914/143903/88/83543/91624/7205/89
GSEA GO和KEGG也是一条命令
gsea.go"BP",OrgDb = org.Hs.eg.db,keyType = "ENTREZID",pvalueCutoff = 1,nPerm = 2)
print(head(gsea.go@result))
## ID Description setSize enrichmentScore NES pvalue p.adjust qvalues rank leading_edge core_enrichment
## GO:0000038 GO:0000038 very long-chain fatty acid metabolic process 25 0.5746498 1.105760 0.3333333 1 1 162 tags=4%, list=0%, signal=4% 641371
## GO:0000183 GO:0000183 chromatin silencing at rDNA 38 0.6412824 1.305810 0.3333333 1 1 5319 tags=29%, list=16%, signal=24% 8368/554313/8357/8350/8358/8354/8364/8366/8361/8362/8363
## GO:0000244 GO:0000244 spliceosomal tri-snRNP complex assembly 23 0.7247442 1.409403 0.3333333 1 1 2298 tags=17%, list=7%, signal=16% 26830/103625684/26832/101954278
## GO:0000353 GO:0000353 formation of quadruple SL/U4/U5/U6 snRNP 10 0.8421648 1.564233 0.3333333 1 1 2298 tags=40%, list=7%, signal=37% 26830/103625684/26832/101954278
## GO:0000365 GO:0000365 mRNA trans splicing, via spliceosome 10 0.8421648 1.564233 0.3333333 1 1 2298 tags=40%, list=7%, signal=37% 26830/103625684/26832/101954278
## GO:0000387 GO:0000387 spliceosomal snRNP assembly 48 0.5683015 1.126044 0.3333333 1 1 2298 tags=10%, list=7%, signal=10% 26830/103625684/26832/6606/101954278
gsea.kegg"hsa",keyType = "kegg",pvalueCutoff = 1,nPerm = 2)
print(head(gsea.kegg@result))
## ID Description setSize enrichmentScore NES pvalue p.adjust qvalues rank leading_edge
## hsa04740 hsa04740 Olfactory transduction 429 0.6026527 1.1857484 0.3333333 1 1 7631 tags=40%, list=23%, signal=31%
## hsa00053 hsa00053 Ascorbate and aldarate metabolism 26 0.5876578 0.9544008 0.6666667 1 1 9375 tags=46%, list=28%, signal=33%
## hsa01040 hsa01040 Biosynthesis of unsaturated fatty acids 27 0.5918780 0.9730072 0.6666667 1 1 162 tags=4%, list=0%, signal=4%
## hsa04061 hsa04061 Viral protein interaction with cytokine and cytokine receptor 97 0.5167078 0.9823899 0.6666667 1 1 8242 tags=34%, list=25%, signal=26%
## hsa04742 hsa04742 Taste transduction 82 0.5508969 1.0035731 0.6666667 1 1 7867 tags=27%, list=24%, signal=21%
## hsa05144 hsa05144 Malaria 49 0.5541224 0.9508375 0.6666667 1 1 10406 tags=45%, list=31%, signal=31%
## core_enrichment
## hsa04740 390538/120787/283162/402135/81050/403244/388761/390063/392391/256892/119678/283297/403282/128360/219479/26735/401993/219493/219432/128367/219431/256148/219958/119694/81327/390077/127059/125962/283189/26737/26219/26682/138804/26716/343173/391107/144124/219477/401667/390260/403274/219952/390142/81309/254786/341799/81318/81466/26188/343170/286365/219873/390078/91860/254973/219437/138803/390148/391196/134083/219957/26530/283110/119765/4992/122748/442184/143496/254783/392309/81469/119687/119695/26246/343169/26538/26492/219983/219956/389090/390261/119692/26532/392138/81392/79346/138883/26338/390442/128368/390892/283111/442185/401663/341416/79501/390084/403257/79541/219870/391121/79324/403277/282775/442191/122742/283159/343172/283092/120776/127074/391192/79290/51764/341568/121275/390037/401666/390093/219968/26693/120066/26740/135946/219428/120065/390326/390036/81472/219869/284433/219981/285659/219482/26212/219453/81797/8387/127385/392390/284383/219487/79490/338675/441911/138799/390167/390082/392392/391109/127062/254879/119682/391195/121130/219959/26595/256144/391191/26494/126541/26245/390152/390067/390191/81697/390649/391194/284532/4994
## hsa00053 7365/10941/54578/54658/10720/54575/54579/54576/54657/54577/54600/79799
## hsa01040 641371
## hsa04061 6351/6366/5473/6846/6364/6363/6375/8743/6368/7124/2833/11009/51554/3558/3606/3576/6356/6346/6362/1436/3627/10563/1234/8740/1236/1232/2829/3559/729230/6357/6369/6348/6355
## hsa04742 50836/259292/50837/50835/259295/50839/259285/170572/259296/2554/259289/51764/259286/338398/50838/9033/259290/259294/50833/50832/2556/3352
## hsa05144 3820/3553/7124/3043/2993/3458/3606/3576/2994/3082/6403/54106/7099/5175/7097/22914/7412/3586/3592/948/6382/3039
这就是GSEA的分析结果了,我这里没有根据P值过滤,因为是随便选择的测试数据,可能显著性都不怎么好!接下来还可以进行可视化展示结果!
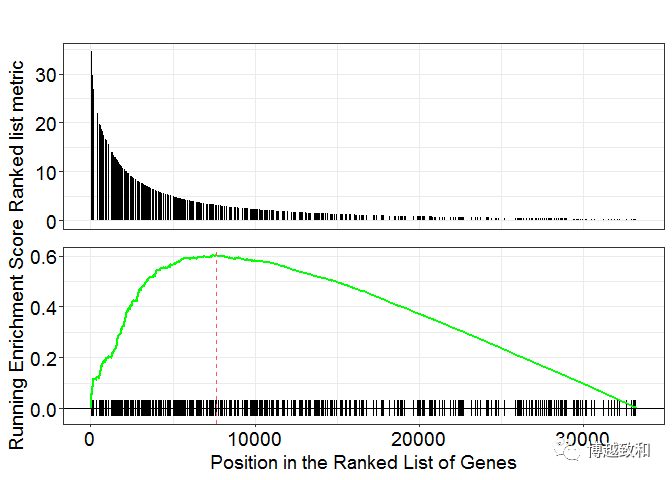
plot 1
gseaplot(gsea.kegg,"hsa04740")
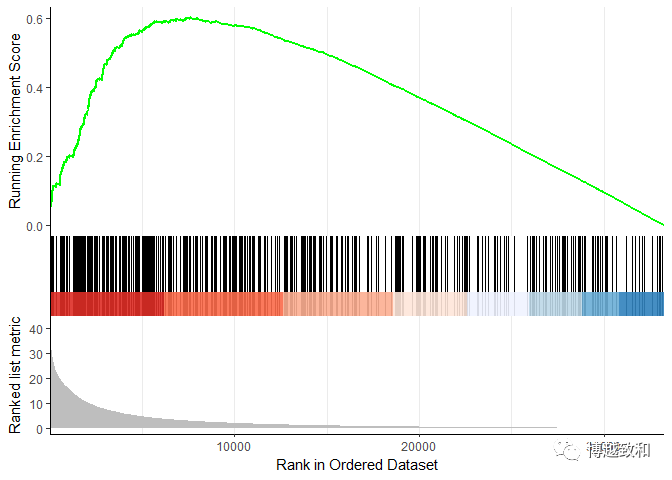
plot 2
library(enrichplot)
enrichplot::gseaplot2(gsea.kegg,"hsa04740")
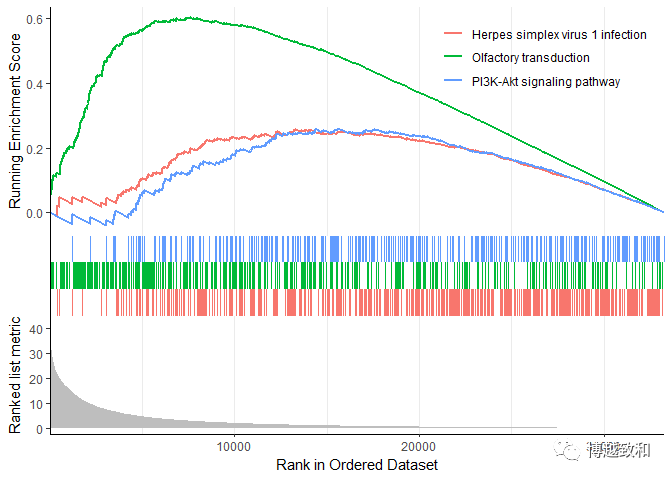
plot 3
enrichplot::gseaplot2(gsea.kegg,c("hsa05168","hsa04740","hsa04151"))
以上就是使用Y叔的clusterProfiler做GSEA分析以及可视化展示的内容!
由于篇幅有限,方法二我们会放到下一期GSEA分析中去,下一期GSEA分析咱们介绍GSEA官网软件 http://software.broadinstitute.org/gsea/index.jsp 来分析并结合TCGA数据和MSigDB基因集,放一张常见的分析图
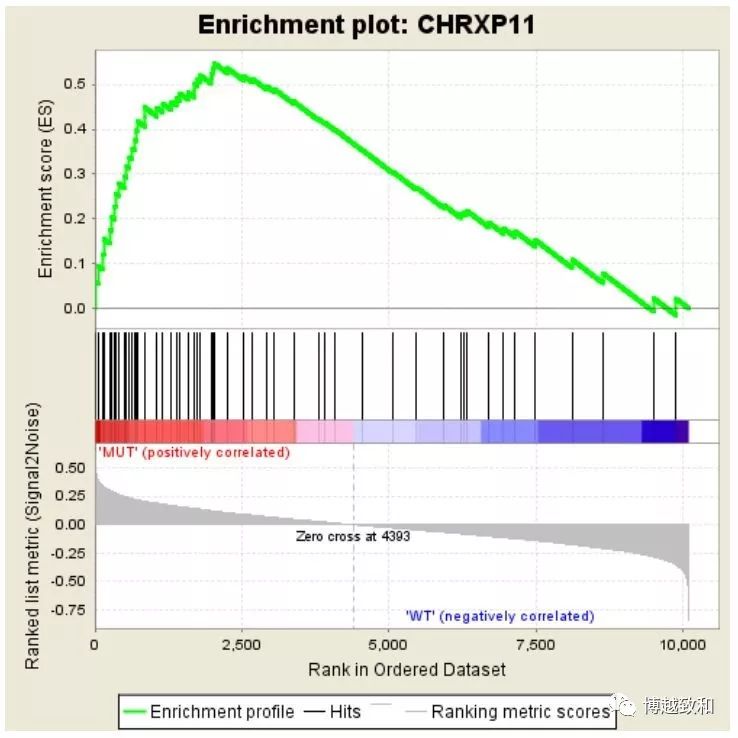
是不是很熟悉,感觉在哪里见过,好了,下一期咱们再见!

参考文献
Guangchuang Yu., Li-Gen Wang, Yanyan Han, Qing-Yu He. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS: A Journal of Integrative Biology. 2012, 16(5):284-287.

扫一扫,了解更多资讯

武汉博越致和生物科技有限公司
个人微信 :hsm664422
电话:027-87705460
您身边的多组学科研助手!