1. 准备工作
硬件:需要配备nvidia独显,RTX3090或RTX2080系列等。较大的显存对分割任务能有一定的帮助。也可以租用谷歌、阿里等服务器,价格一般按提供的使用时间和算力计算。
软件:一般使用ubuntu,借助anaconda配置pytorch环境。IDE个人推荐使用Pycharm。
https://pytorch.org/get-started/locally/
2. 数据预处理
2.1 package
一般的二维图像处理依赖opencv,对于医学数据,如MRI、CT等,可借助SimpleITK,NiBabel等包进行处理。
2.2 基本处理
医学图像文件后缀名一般为nii,nii.gz,dicom等,例举使用simpleitk和opncv的相关基本操作
import SimpleITK as sitk
import cv2
img = sitk.ReadImage(img_filename)
img_array = sitk.GetArrayFromImage(img)
img_array = np.transpose(img_array, (1, 2, 0)) # x,y,z--210 120-yxz
spacing = img.GetSpacing()
origin = img.GetOrigin()
direction = img.GetDirection()
reimg.SetOrigin(origin)
reimg.SetSpacing(tar_spacing)
reimg.SetDirection(direction)
sitk.WriteImage(relab, join(preproc_path, '%s_prelab.nrrd') % filename_now)
# normalization
def norm(img_array):
mri_max = np.amax(img_array)
mri_min = np.amin(img_array)
mri_img = ((img_array - mri_min) / (mri_max - mri_min)) * 255
mri_img = mri_img.astype('uint8')
return mri_img
# contrast limited adaptive histogram equalization
def clahe(mri_img):
h, w, d = mri_img.shape
img_clahe_add = np.zeros_like(mri_img)
for k in range(d):
temp = mri_img[:, :, k]
clahe = cv2.createCLAHE(clipLimit=2.0, tileGridSize=(8, 8))
img_clahe = clahe.apply(temp)
# cv2.imshow('mri', np.concatenate([temp,img_clahe], 1))
# cv2.waitKey(1)
img_clahe_add[:, :, k] = img_clahe
return img_clahe_add3. 训练测试集划分
对于公开数据集,一般已经固定了训练集、验证集和测试集。对于私有数据集,则需要自己进行划分,一般采用交叉验证的方式以说明模型的范化性。
例如k折交叉验证(K-fold cross validation),就是把样本集S分成k份,分别使用其中的(k-1)份作为训练集,剩下的1份作为交叉验证集,最后取最后的平均误差,来评估这个模型。
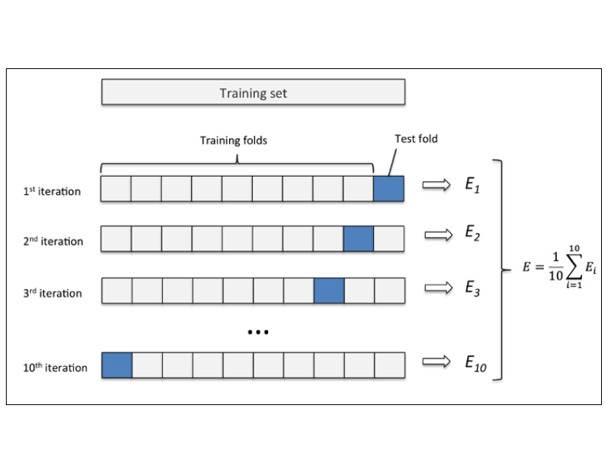
4. 评价指标
每一词训练得到一个输出label,和医生手工标注的金标准进行比较,计算损失并反向传播,不断重复 。
医学上常用的指标为Dice系数,Dice系数是一种集合相似度度量函数,通常用于计算两个样本的相似度,取值范围在[0,1]:
Dice:

其中 |X∩Y| 是X和Y之间的交集,|X|和|Y|分表表示X和Y的元素的个数,其中,分子的系数为2,是因为分母存在重复计算X和Y之间的共同元素的原因。
Dice Loss:
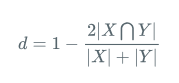
https://zhuanlan.zhihu.com/p/86704421
5. 代码实现
工程实现引用"Attention U-Net: Learning Where to Look for the Pancreas", MIDL'18, Amsterdam
https://github.com/ozan-oktay/Attention-Gated-Networks
5.1 参数配置
涉及大量参数时,可以使用json、yaml等文件记录需要配置的参数,并在主函数入口进行读取。
如下yaml格式文件,保存需要的变量参数
manual_seed: 0
device:
cuda: 0
data:
data_path: data/resampled0302
aug:
zoom_shape: [160,160,32]
shift_val: [0.1,0.1]
rotate_val: 5.0
scale_val: [1.0,1.0]
random_flip_prob: 0.5
train:
is_train: True
is_test: True
n_epochs: 1200
batch_size: 1
model:
task: segment
model_name: matt
criterion: dice_ce
in_channels: 1
n_classes: 2
optimizer: adam
feature_scale: 8在初始化时,读取config并为相应变量赋值
class mainSegment:
def __init__(self, config):
self.timestamp = "{0:%Y%m%d_T%H.%M.%S/}".format(datetime.now())
self.timestamp_y = "{0:%Y%m%d/}".format(datetime.now())
# get config from yaml file
self.path_config = config
config = load_config(config)
self.config = config
self.data_path = self.config['data'].get('data_path')
self.zoom_shape = self.config['aug'].get('zoom_shape')
self.train_Opts = self.config.get('train')
self.aug_Opts = self.config.get('aug')
self.model_Opts = self.config.get('model')
self.n_classes = self.model_Opts.get('n_classes')
logger.info(config)5.2 模型读取
# model init
if self.model_Opts.get('model_name') == 'unet3d_myedit':
self.model = Unet3d_myedit(self.model_Opts).cuda()
if self.model_Opts.get('model_name') == 'smallHRNet':
self.model = getSmallHRNet(NoLabels=2).cuda()5.3 数据准备
数据的读入可重写pytorch提供的torch.utils.data.dataset,使用torch.utils.data.Dataloader,在每一个epoch中将数据读取并进行数据增强,喂给模型。
get_aug = get_augmentation(self.aug_Opts)
train_dataset = SegDataset(self.data_path,
split='train',
zoom_shape=self.zoom_shape,
transform=get_aug['train'])
test_dataset = SegDataset(self.data_path,
split='test',
zoom_shape=self.zoom_shape,
transform=get_aug['test'])
train_loader = DataLoader(dataset=train_dataset,
batch_size=self.train_Opts.get('batch_size', 1),
shuffle=True,
num_workers=0
)
test_loader = DataLoader(dataset=test_dataset,
batch_size=1,
shuffle=False,
num_workers=0)
Dataset类的重写如下:
class SegDataset(data.Dataset):
def __init__(self, root_dir, split, zoom_shape, transform=None):
# get parameters
self.root_dir = root_dir
self.split = split
self.zoom_shape = zoom_shape
self.transform = transform
self.image_dir = join(root_dir, split, 'augimgnpy')
self.target_dir = join(root_dir, split, 'auglabnpy')
self.filenames = sorted(listdir(self.image_dir))
self.image_filenames = sorted([join(self.image_dir, x) for x in listdir(self.image_dir)]) # 01 02
self.target_filenames = sorted([join(self.target_dir, x) for x in listdir(self.target_dir)])
def __len__(self):
return len(self.image_filenames)
def __getitem__(self, index):
img_arr = np.load(self.image_filenames[index]).astype(np.float32)
target = np.load(self.target_filenames[index]).astype(np.uint8)
name_now = self.filenames[index][:-11]
img_shape = np.array(img_arr.shape)
target_shape = np.array(target.shape)
img_arr = scipy.ndimage.zoom(img_arr, np.array(self.zoom_shape) / img_shape, order=0).astype(np.float32)
target = scipy.ndimage.zoom(target, np.array(self.zoom_shape) / target_shape, order=0).astype(np.uint8)
# process the label for different organs
if self.transform:
img_arr, target = self.transform(img_arr, target)
return img_arr, target, name_now
数据增强,包括随机翻转,随机旋转,随机平移等操作。依赖参考开源库中的torchsample实现,代码如下:
from torchsample import transforms as ts
def get_augmentation(opts=None):
trans_obj = seg_Transform()
if opts:
trans_obj.initialize(opts)
trans_obj.print()
return trans_obj.augmentation()
class seg_Transform:
def __init__(self,):
# Affine and Intensity Transformations
self.shift_val = (0.1, 0.1)
self.rotate_val = 5.0
self.scale_val = (1.0, 1.0)
self.random_flip_prob = 0.5
def print(self):
print('\n\n############# Augmentation Parameters #############')
pprint(vars(self))
print('###################################################\n\n')
def initialize(self, opts):
t_opts = opts
if 'random_flip_prob' in t_opts.keys():
self.random_flip_prob = t_opts.get('random_flip_prob')
def augmentation(self):
train_transform = ts.Compose([
ts.ToTensor(),
ts.ChannelsFirst(),
ts.TypeCast(['float', 'float']),
ts.RandomFlip(h=True, v=True, p=self.random_flip_prob),
ts.RandomAffine(rotation_range=self.rotate_val, translation_range=self.shift_val,
zoom_range=self.scale_val, interp=('bilinear', 'nearest')),
ts.ChannelsLast(),
ts.AddChannel(axis=0),
ts.TypeCast(['float', 'long'])
])
test_transform = ts.Compose([
ts.ToTensor(),
ts.ChannelsFirst(),
ts.TypeCast(['float', 'float']),
# #
# #
ts.ChannelsLast(),
ts.AddChannel(axis=0),
ts.TypeCast(['float', 'long'])
])
return {'train': train_transform, 'test': test_transform}
5.4 训练过程
核心代码段,包括tensorboard的使用,配置文件的备份保存。
if self.train_Opts.get('is_train'):
total_epoch = self.train_Opts.get('n_epochs')
lowest_loss = 10
# setup tensorboardX
writer = SummaryWriter(self.save_path)
for epoch in range(total_epoch):
epoch_loss = 0
epoch_dice1 = 0
for epoch_iter, (images, labels, name_now) in tqdm(enumerate(train_loader, 1), total=len(train_loader)):
images, labels, name_now = images.cuda(), labels.cuda(), name_now
self.optimizer.zero_grad()
outputs = self.model(images)
loss = self.criterion(outputs, labels)
epoch_loss += loss.item()
loss.backward()
self.optimizer.step()
_, _, _, dice_score, _, _ = segmentation_stats(outputs, labels)
if self.n_classes == 2:
epoch_dice1 += dice_score[1]
# print(epoch_iter)
if self.n_classes == 2:
print("Epoch [%d/%d], train_Loss: %.4f, dice_1: %.4f"
% (epoch + 1, total_epoch, epoch_loss / epoch_iter, epoch_dice1 / epoch_iter))
# save model
torch.save(self.model.state_dict(), os.path.join(self.save_path, self.model_file))
# save best model (lowest loss)
if epoch_loss/epoch_iter <= lowest_loss:
torch.save(self.model.state_dict(), os.path.join(self.save_path, self.model_file_best))
lowest_loss = epoch_loss/epoch_iter
# test
test_dice1 = 0
if self.n_classes == 2:
test_dice1 = self.test(self.model, test_loader, self.model_file, is_visualize=False)
# visualize using tensorboard
writer.add_scalar('data/train_loss', epoch_loss/epoch_iter, epoch)
writer.add_scalar('data/train_dice_1', epoch_dice1/epoch_iter, epoch)
writer.add_scalar('data/test_dice_1', test_dice1, epoch)
writer.close()
# model_file_path = os.path.join(os.getcwd(), model_file)
# model_file_best_path = os.path.join(os.getcwd(), model_file_best)
config_path = os.path.join(os.getcwd(), self.path_config)
# shutil.copy(model_file_path, save_path)
# shutil.copy(model_file_best_path, save_path)
shutil.copy(config_path, self.save_path)
print_hi('PyCharm')5.5 测试过程
def test(self, model, test_loader, model_file, is_visualize=False):
model.load_state_dict(torch.load(os.path.join(self.save_path, model_file)))
model.eval()
total_dice1 = 0
for iter, (images, labels, name_now) in tqdm(enumerate(test_loader, 1), total=len(test_loader)):
images, labels, name_now = images.cuda(), labels.cuda(), name_now
prediction = model(images)
overall_acc, mean_acc, mean_iou, dice_score, _, _ = segmentation_stats(prediction, labels)
if self.n_classes == 2:
total_dice1 += dice_score[1]
print(name_now, "test_dice1:%.4f " % (dice_score[1]))
if self.n_classes == 2:
print("ave test_dice1:%.4f " % (total_dice1 / len(test_loader)))
return total_dice1 / len(test_loader)5.6 模型实现
以实现经典的Unet网络为例,其结构如下所示:
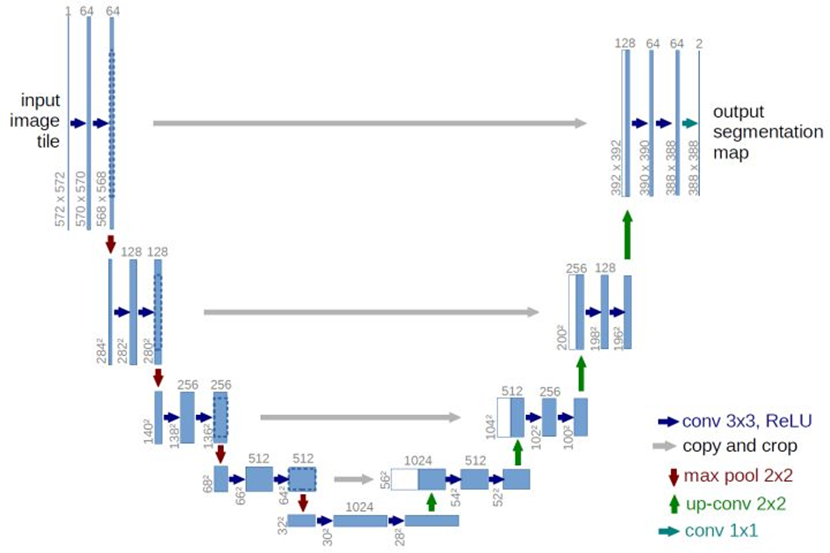
Ronneberger O, Fischer P, Brox T. U-net: Convolutional networks for biomedical image segmentation[C]//International Conference on Medical image computing and computer-assisted intervention. Springer, Cham, 2015: 234-241.
代码如下:
import torch
from torch import nn
import torch.nn.functional as F
from collects.utils import init_weights
class myUnet3D_2(nn.Module):
def __init__(self, config):
super(myUnet3D_2, self).__init__()
self.in_channels = config.get('in_channels')
self.n_classes = config.get('n_classes')
self.feature_scale = config.get('feature_scale')
self.is_deconv = True
filters = [64, 128, 256, 512, 1024]
filters = [int(x / self.feature_scale) for x in filters]
# downsampling
self.conv1 = UnetConv3(self.in_channels, filters[0])
self.maxpool1 = nn.MaxPool3d(kernel_size=(2, 2, 1))
self.conv2 = UnetConv3(filters[0], filters[1])
self.maxpool2 = nn.MaxPool3d(kernel_size=(2, 2, 1))
self.conv3 = UnetConv3(filters[1], filters[2])
self.maxpool3 = nn.MaxPool3d(kernel_size=(2, 2, 1))
self.conv4 = UnetConv3(filters[2], filters[3])
self.maxpool4 = nn.MaxPool3d(kernel_size=(2, 2, 1))
self.center = UnetConv3(filters[3], filters[4])
# upsampling
self.up_concat4 = UnetUp3(filters[4], filters[3])
self.up_concat3 = UnetUp3(filters[3], filters[2])
self.up_concat2 = UnetUp3(filters[2], filters[1])
self.up_concat1 = UnetUp3(filters[1], filters[0])
# final conv (without any concat)
self.final = nn.Conv3d(filters[0], self.n_classes, 1)
# initialise weights
for m in self.modules():
if isinstance(m, nn.Conv3d):
init_weights(m, init_type='kaiming')
elif isinstance(m, nn.GroupNorm):
init_weights(m, init_type='kaiming')
def forward(self, inputs):
conv1 = self.conv1(inputs)
maxpool1 = self.maxpool1(conv1)
conv2 = self.conv2(maxpool1)
maxpool2 = self.maxpool2(conv2)
conv3 = self.conv3(maxpool2)
maxpool3 = self.maxpool3(conv3)
conv4 = self.conv4(maxpool3)
maxpool4 = self.maxpool4(conv4)
center = self.center(maxpool4)
up4 = self.up_concat4(conv4, center)
up3 = self.up_concat3(conv3, up4)
up2 = self.up_concat2(conv2, up3)
up1 = self.up_concat1(conv1, up2)
final = self.final(up1)
return final
class UnetConv3(nn.Module):
def __init__(self, in_size, out_size, kernel_size=(3, 3, 1), padding_size=(1, 1, 0), init_stride=(1, 1, 1)):
super(UnetConv3, self).__init__()
self.conv1 = nn.Sequential(nn.Conv3d(in_size, out_size, kernel_size, init_stride, padding_size),
nn.GroupNorm(num_groups=2, num_channels=out_size),
nn.ReLU(inplace=False))
self.conv2 = nn.Sequential(nn.Conv3d(out_size, out_size, kernel_size, 1, padding_size),
nn.GroupNorm(num_groups=2, num_channels=out_size),
nn.ReLU(inplace=False))
# initialise the blocks
for m in self.children():
init_weights(m, init_type='kaiming')
def forward(self, inputs):
outputs = self.conv1(inputs)
outputs = self.conv2(outputs)
return outputs
class UnetUp3(nn.Module):
def __init__(self, in_size, out_size):
super(UnetUp3, self).__init__()
self.conv = UnetConv3(in_size, out_size)
self.up = nn.ConvTranspose3d(in_size, out_size, kernel_size=(4, 4, 1), stride=(2, 2, 1), padding=(1, 1, 0))
# self.up = nn.Sequential(nn.Conv3d(in_size, out_size, kernel_size=(1, 1, 1)),
# nn.Upsample(scale_factor=(2, 2, 1), mode='nearest'))
# self.up = nn.Upsample(scale_factor=(2, 2, 1), mode='trilinear')
# initialise the blocks
for m in self.children():
if m.__class__.__name__.find('UnetConv3') != -1: continue
init_weights(m, init_type='kaiming')
def forward(self, inputs1, inputs2):
outputs2 = self.up(inputs2)
offset = outputs2.size()[2] - inputs1.size()[2]
padding = 2 * [offset // 2, offset // 2, 0]
outputs1 = F.pad(inputs1, padding)
return self.conv(torch.cat([outputs1, outputs2], 1))