预处理和主成分分析(PCA)
目的:
- 学会对数据进行预处理与缩放
- 学会使用PCA进行降维
- 初步了解相关参数
要求:
基于乳腺癌数据集完成以下任务:
1、乳腺癌数据的特征值有多少?
2、写出数据变换的一般步骤
3、将数据进行StandardScale,MinMaxScaler和RobustScaer转换
4、在第三步骤的基础上,选择两个主成分,三个主成分利用LinearSVC分别器,求不同情况下的精度(测试集的精度)。
因为考虑到每次切片的random_state都不同,为了使测试精度更有说服力,每次给出的精度都经过n次切片训练,n是自己设定的。
1、 乳腺癌数据的特征值
#获取数据集
cancer = load_breast_cancer()
print(cancer.data.shape)
可以看到输出结果:
所以乳腺癌数据有30个特征
2、 数据变换的一般步骤:
1) 建立模型(以StandardScale为例) st_Scaler = StandardScaler()
2) 训练模型 st_Scaler.fit(cancer.data)
3) 数据转换 cancer_stScaler = st_Scaler.transform(cancer.data)
4、 在第三步骤的基础上,选择两个主成分,三个主成分利用LinearSVC分别器,求不同情况下的精度(测试集的精度)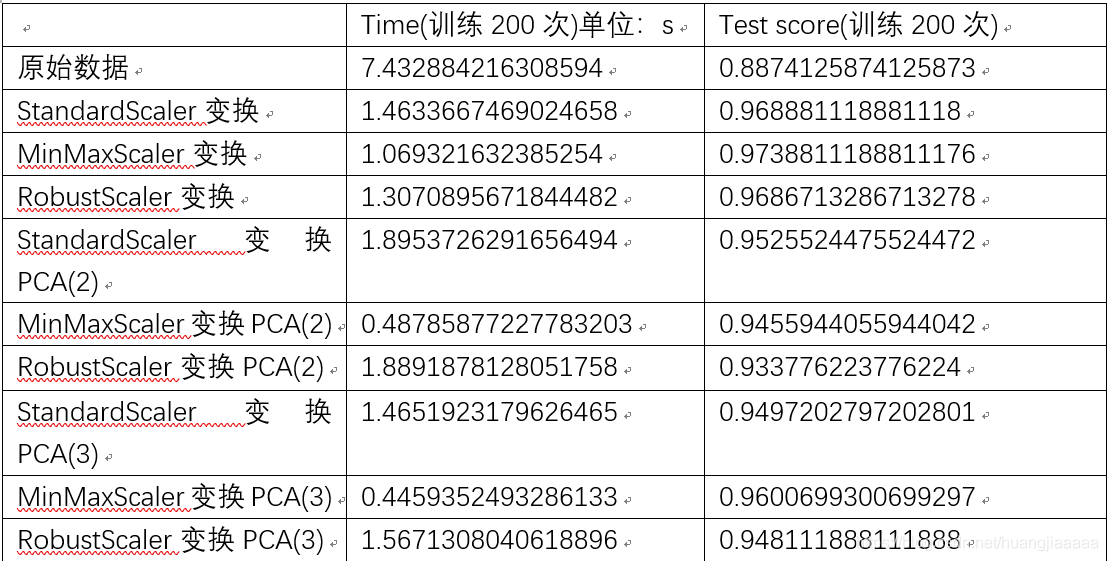
基于上表,我们可以看出数据经过变换处理后,训练速度有明显的提升,精度也提升了,所以可以看出对数据的处理是非常必要的,不仅能提升时间性能,还能提升测试的精度。PCA降维是为了降低分析的难度,当一个数据集的特征非常非常多的时候,我们通常要很费时间的去训练,但是通过上表我们可以看出,这个数据集原本是30维,降到2维以后,测试精度也并没下降,反而有所上升,所以我们认为,当数据集有大量的特征,进行PCA降维是有必要的。
完整代码:
# -*- coding: utf-8 -*-
"""
Created on Fri Nov 15 20:20:17 2019
@author: asus
"""
"""
预处理和主成分分析(PCA)
目的:
1. 学会对数据进行预处理与缩放
2. 学会使用PCA进行降维
3. 初步了解相关参数
要求:
基于乳腺癌数据集完成以下任务:
1、乳腺癌数据的特征值有多少?
2、写出数据变换的一般步骤
3、将数据进行StandardScale,MixMaxScaler和RobustScaer转换
4、在第三步骤的基础上,选择两个主成分,三个主成分利用LinearSVC分别器,求不同情况下的精度(测试集的精度)。
#注意
数据集使用
from sklearn.datasets import load_breast_cancer
cancer = load_breast_cancer()
"""
import mglearn
import matplotlib.pyplot as plt
import numpy as np
from sklearn.datasets import load_breast_cancer
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn.decomposition import PCA
from sklearn.preprocessing import MinMaxScaler
from sklearn.preprocessing import RobustScaler
from sklearn.preprocessing import Normalizer
from sklearn.svm import LinearSVC
import time
import warnings
warnings.filterwarnings("ignore")#消除警告
def Linear_svc_mean_Score(data_set,target_set,num):
"""
来一组数据,训练num次,返回linear_svc的训练集与测试集的平均得分
入口参数为数据集,目标数据集,训练次数
"""
train_scores = []
test_scores = []
start = time.time()
for i in range(num):
X_train,X_test,y_train,y_test = train_test_split(data_set,target_set,stratify = target_set)
linear_svc.fit(X_train,y_train)
train_score = linear_svc.score(X_train, y_train)
test_score = linear_svc.score(X_test, y_test)
train_scores.append(train_score)
test_scores.append(test_score)
print("Time used :",time.time() - start)
train_mean_scores = sum(train_scores) / len(train_scores)
test_mean_scores = sum(test_scores) / len(test_scores)
return (train_mean_scores,test_mean_scores)
#获取数据集
cancer = load_breast_cancer()
print(cancer.data.shape)
print(cancer.target.shape)
#对原始数据切片
X_train,X_test,y_train,y_test = train_test_split(cancer.data,cancer.target,stratify = cancer.target,random_state = 0)
#建立模型
linear_svc = LinearSVC()
#训练模型
linear_svc.fit(X_train,y_train)
#对原始数据训练队的模型进行评估
#print("The train scores of Raw data :",linear_svc.score(X_train,y_train))
#print("The test scores of Raw data :",linear_svc.score(X_test,y_test))
#print("Raw data : ",X_train.shape,X_train[0])
#训练60次
num = 200
train_mean_scores,test_mean_scores = Linear_svc_mean_Score(cancer.data,cancer.target,num)
#print("The train scores of Raw data :",train_mean_scores)
print("The test scores of Raw data :",test_mean_scores)
#将数据进行StandardScale变换
st_Scaler = StandardScaler()
st_Scaler.fit(cancer.data)
cancer_stScaler = st_Scaler.transform(cancer.data)
train_mean_scores,test_mean_scores = Linear_svc_mean_Score(cancer_stScaler,cancer.target,num)
#print("The train scores of StandardScaler transformed data :",train_mean_scores)
print("The test scores of StandardScaler transformed data :",test_mean_scores)
#在StandardScale基础上,选择两个主成分
pca = PCA(n_components = 3)
pca.fit(cancer_stScaler)
X_pca = pca.transform(cancer_stScaler)
X_train,X_test,y_train,y_test = train_test_split(X_pca,cancer.target,stratify=cancer.target,random_state=60)
#查看PCA降维后的数据类型
print("PCA(2) data : " ,X_train.shape,X_train[0])
train_mean_scores,test_mean_scores = Linear_svc_mean_Score(X_pca,cancer.target,num)
#print("The train scores of StandardScaler transformed(PCA(2)) data :",train_mean_scores)
print("The test scores of StandardScaler transformed(PCA(2)) data :",test_mean_scores)
#将数据进行MixMaxScaler变换
M_scaler = MinMaxScaler()
M_scaler.fit(cancer.data)
cancer_M_Scaler = M_scaler.transform(cancer.data)
train_mean_scores,test_mean_scores = Linear_svc_mean_Score(cancer_M_Scaler,cancer.target,num)
#print("The train scores of MinMaxScaler transformed data :",train_mean_scores)
print("The test scores of MinMaxScaler transformed data :",test_mean_scores)
#在MixMaxScaler基础上,选择两个主成分
pca = PCA(n_components = 3)
pca.fit(cancer_M_Scaler)
X_pca = pca.transform(cancer_M_Scaler)
X_train,X_test,y_train,y_test = train_test_split(X_pca,cancer.target,stratify=cancer.target,random_state=60)
#查看PCA降维后的数据类型
print("PCA(2) data : " ,X_train.shape,X_train[0])
train_mean_scores,test_mean_scores = Linear_svc_mean_Score(X_pca,cancer.target,num)
#print("The train scores of MinMaxScaler transformed(PCA(2)) data :",train_mean_scores)
print("The test scores of MinMaxScaler transformed(PCA(2)) data :",test_mean_scores)
#在RobustScaler基础上,选择两个主成分
R_scaler = RobustScaler()
R_scaler.fit(cancer.data)
cancer_R_Scaler = R_scaler.transform(cancer.data)
train_mean_scores,test_mean_scores = Linear_svc_mean_Score(cancer_R_Scaler,cancer.target,num)
#print("The train scores of RobustScaler transformed data :",train_mean_scores)
print("The test scores of RobustScaler transformed data :",test_mean_scores)
#在RobustScaler基础上,选择两个主成分
pca = PCA(n_components = 3)
pca.fit(cancer_R_Scaler)
X_pca = pca.transform(cancer_R_Scaler)
X_train,X_test,y_train,y_test = train_test_split(X_pca,cancer.target,stratify=cancer.target,random_state=60)
#查看PCA降维后的数据类型
print("PCA(2) data : " ,X_train.shape,X_train[0])
train_mean_scores,test_mean_scores = Linear_svc_mean_Score(X_pca,cancer.target,num)
#print("The train scores of RobustScaler transformed(PCA(2)) data :",train_mean_scores)
print("The test scores of RobustScaler transformed(PCA(2)) data :",test_mean_scores)
参考书籍:《python机器学习基础教程》
版权声明:本文为huangjiaaaaa原创文章,遵循CC 4.0 BY-SA版权协议,转载请附上原文出处链接和本声明。